Denoising ADT#
In this tutorial, we go through steps for ADT denoising in CITE-seq.
We will use the 10xPBMC5k CITE-seq dataset, which contained about five thousands cells stained with a panel of 32 antibodies. The original dataset was downloaded from 10x genomics dataset, and the processed and annotation data is available at scAR-reproducibility/data
[ ]:
# Run this cell to install scar in Colab
# Skip this cell if running on your own device
%pip install scanpy
%pip install git+https://github.com/Novartis/scAR.git
%pip install matplotlib==3.1.3 # Specify this matplotlib version to avoid errors
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scanpy as sc
from scar import model
import warnings
warnings.simplefilter("ignore")
Matplotlib is building the font cache; this may take a moment.
Load data#
Cells were annotated using well-established markers, see the scAR manuscript for details. For the tutoring purpose, the processed file (CITEseq_PBMCs_5k.h5ad) is provided.
[2]:
!git clone https://github.com/CaibinSh/scAR-reproducibility.git
PBMCs5k = sc.read('scAR-reproducibility/data/CITEseq_PBMCs_5k.h5ad')
fatal: destination path 'scAR-reproducibility' already exists and is not an empty directory.
visualization of cell types
[3]:
sc.settings.set_figure_params(dpi=150,figsize = (3, 3.1))
sc.pl.umap(PBMCs5k, size=10, color=["subtype"],color_map="viridis",legend_loc="on data", frameon=False, legend_fontsize=5)

Raw count matrix#
Get a dataframe of ADT raw count as the input of scar
[4]:
raw_ADT = PBMCs5k[:, PBMCs5k.var_names.str.endswith('_TotalSeqB')].to_df()
raw_ADT.columns = raw_ADT.columns.str.replace('_TotalSeqB','')
raw_ADT.head()
[4]:
| CD3 | CD4 | CD8a | CD11b | CD14 | CD15 | CD16 | CD19 | CD20 | CD25 | ... | CD197 | CD274 | CD278 | CD335 | PD-1 | HLA-DR | TIGIT | IgG1_control | IgG2a_control | IgG2b_control | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCGGGTAGTTCAAACC-1 | 876.0 | 968.0 | 8.0 | 10.0 | 8.0 | 9.0 | 1.0 | 3.0 | 10.0 | 3.0 | ... | 20.0 | 2.0 | 29.0 | 6.0 | 6.0 | 14.0 | 4.0 | 3.0 | 3.0 | 2.0 |
| CTTTCGGTCCGCAACG-1 | 516.0 | 664.0 | 4.0 | 4.0 | 4.0 | 6.0 | 0.0 | 3.0 | 1.0 | 2.0 | ... | 15.0 | 1.0 | 26.0 | 5.0 | 5.0 | 8.0 | 1.0 | 1.0 | 3.0 | 0.0 |
| TCTACCGGTGTAAACA-1 | 666.0 | 837.0 | 5.0 | 10.0 | 6.0 | 5.0 | 1.0 | 4.0 | 7.0 | 4.0 | ... | 30.0 | 1.0 | 36.0 | 2.0 | 5.0 | 9.0 | 1.0 | 3.0 | 3.0 | 2.0 |
| ATAGACCCATAATGCC-1 | 352.0 | 709.0 | 3.0 | 7.0 | 8.0 | 12.0 | 3.0 | 4.0 | 4.0 | 2.0 | ... | 13.0 | 2.0 | 100.0 | 6.0 | 12.0 | 9.0 | 4.0 | 2.0 | 2.0 | 2.0 |
| TCTATACAGGGTATAT-1 | 289.0 | 861.0 | 7.0 | 3.0 | 8.0 | 4.0 | 2.0 | 4.0 | 9.0 | 21.0 | ... | 10.0 | 3.0 | 31.0 | 3.0 | 3.0 | 11.0 | 1.0 | 4.0 | 4.0 | 4.0 |
5 rows × 32 columns
Ambient profile#
Get a dataframe of ADT ambient profile as the input of scar
[5]:
ambient_profile = PBMCs5k.uns['ambient_profile']
ambient_profile.head()
[5]:
| ambient_profile | |
|---|---|
| CD3 | 0.049876 |
| CD4 | 0.048823 |
| CD8a | 0.023453 |
| CD11b | 0.083170 |
| CD14 | 0.107178 |
IMPORTANT
In ‘CITEseq’ mode, we do not recommend running scar using default setting (i.e. estimating ambient profile by averaging cells). Instead, we recommend calculating ambient profile by averaging cell-free droplets. Calculation of ambient profile is key to precise denoising.
See an example for calculating ambient profile.
Training#
[6]:
ADT_scar = model(raw_count = raw_ADT,
ambient_profile = ambient_profile, # Providing ambient profile is recommended for CITEseq; in other modes, you can leave this argument as the default value -- None
feature_type = 'ADT', # "ADT" or "ADTs" for denoising protein counts in CITE-seq
count_model = 'binomial', # Depending on your data's sparsity, you can choose between 'binomial', 'possion', and 'zeroinflatedpossion'
)
ADT_scar.train(epochs=500,
batch_size=64,
verbose=True
)
# After training, we can infer the true protein signal
ADT_scar.inference() # by defaut, batch_size=None, set a batch_size if getting GPU memory issue
2023-05-01 16:29:45|INFO|model|No GPU detected. Use CPU instead.
2023-05-01 16:29:47|INFO|VAE|Running VAE using the following param set:
2023-05-01 16:29:47|INFO|VAE|...denoised count type: ADT
2023-05-01 16:29:47|INFO|VAE|...count model: binomial
2023-05-01 16:29:47|INFO|VAE|...num_input_feature: 32
2023-05-01 16:29:47|INFO|VAE|...NN_layer1: 150
2023-05-01 16:29:47|INFO|VAE|...NN_layer2: 100
2023-05-01 16:29:47|INFO|VAE|...latent_space: 15
2023-05-01 16:29:47|INFO|VAE|...dropout_prob: 0.00
2023-05-01 16:29:47|INFO|VAE|...expected data sparsity: 0.90
2023-05-01 16:29:47|INFO|model|kld_weight: 1.00e-05
2023-05-01 16:29:47|INFO|model|learning rate: 1.00e-03
2023-05-01 16:29:47|INFO|model|lr_step_size: 5
2023-05-01 16:29:47|INFO|model|lr_gamma: 0.97
Training: 100%|██████████| 500/500 [02:52<00:00, 2.90it/s, Loss=1.0789e+02]
The denoised counts are saved in ADT_scar.native_counts and can be used for downstream analysis.
[7]:
denoised_ADT = pd.DataFrame(ADT_scar.native_counts, index=raw_ADT.index, columns=raw_ADT.columns)
denoised_ADT.head()
[7]:
| CD3 | CD4 | CD8a | CD11b | CD14 | CD15 | CD16 | CD19 | CD20 | CD25 | ... | CD197 | CD274 | CD278 | CD335 | PD-1 | HLA-DR | TIGIT | IgG1_control | IgG2a_control | IgG2b_control | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCGGGTAGTTCAAACC-1 | 865.0 | 973.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 9.0 | 0.0 | 26.0 | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| CTTTCGGTCCGCAACG-1 | 502.0 | 648.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 7.0 | 0.0 | 23.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| TCTACCGGTGTAAACA-1 | 660.0 | 855.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 2.0 | 0.0 | ... | 9.0 | 0.0 | 34.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ATAGACCCATAATGCC-1 | 362.0 | 706.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 5.0 | 0.0 | 97.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| TCTATACAGGGTATAT-1 | 292.0 | 870.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 1.0 | 21.0 | ... | 6.0 | 0.0 | 26.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
5 rows × 32 columns
Visualization#
Plot setting
[8]:
from matplotlib import pylab
params = {'legend.fontsize': 6,
'legend.title_fontsize': 8,
'figure.facecolor':"w",
'figure.figsize': (6, 4.5),
'axes.labelsize': 6,
'axes.titlesize':8,
'axes.linewidth': 0.5,
'xtick.labelsize':6,
'ytick.labelsize':6,
'axes.grid':False,}
pylab.rc('font',**{'family':'serif','serif':['Palatino'],'size':10})
pylab.rcParams.update(params);
sns.set_palette("muted");
sns.set_style("ticks");
sns.despine(offset=4, trim=True);
<Figure size 900x675 with 0 Axes>
Boxplot of ADT counts per cell type#
Prepare a dataframe for visualization
[9]:
denoised_ADT_stacked = np.log2(denoised_ADT+1).stack().to_frame('log2(ADT+1)').rename_axis(['cells', 'ADTs']).reset_index().set_index('cells')
denoised_ADT_stacked = denoised_ADT_stacked.join(PBMCs5k.obs)
denoised_ADT_stacked['count'] = 'scar-denoised ADT'
raw_ADT_stacked = np.log2(raw_ADT+1).stack().to_frame('log2(ADT+1)').rename_axis(['cells', 'ADTs']).reset_index().set_index('cells') # log scale for visualization,
raw_ADT_stacked = raw_ADT_stacked.join(PBMCs5k.obs) # Add cell type information
raw_ADT_stacked['count'] = 'raw ADT'
ADTs_stacked = raw_ADT_stacked.append(denoised_ADT_stacked)
ADTs_stacked.head()
[9]:
| ADTs | log2(ADT+1) | celltype | subtype | count | |
|---|---|---|---|---|---|
| AAACCCAAGAGACAAG-1 | CD3 | 4.700440 | Monocytes | intermediate mono | raw ADT |
| AAACCCAAGAGACAAG-1 | CD4 | 7.366322 | Monocytes | intermediate mono | raw ADT |
| AAACCCAAGAGACAAG-1 | CD8a | 4.087463 | Monocytes | intermediate mono | raw ADT |
| AAACCCAAGAGACAAG-1 | CD11b | 11.556027 | Monocytes | intermediate mono | raw ADT |
| AAACCCAAGAGACAAG-1 | CD14 | 9.445015 | Monocytes | intermediate mono | raw ADT |
Boxplot visualization of ADT counts per cell type before and after denoising.
[10]:
marker = "CD19"
tmp = ADTs_stacked[ADTs_stacked['ADTs']==marker]
plt.figure(figsize=(3, 1.8))
g = sns.boxplot(x="subtype",
y="log2(ADT+1)",
data=tmp,
hue="count",
hue_order=['raw ADT', 'scar-denoised ADT'],
orient="v",
dodge=True,
width=0.8,
showfliers=False,
linewidth=0.5,
);
g.legend_.set_title('')
g.set(xlabel='',title=f'{marker} protein counts');
g.set_xticklabels(g.get_xticklabels(),rotation=45, ha='right');
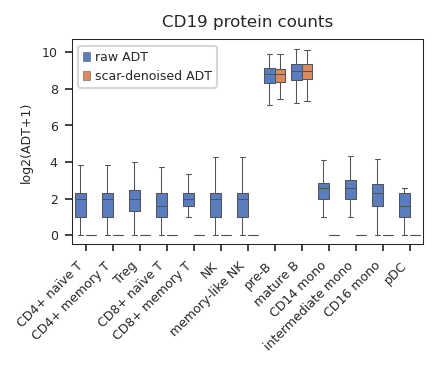
CD19 is a B cell specific marker, raw CD19 counts are detected in every cell type, while denoised counts are only observed in B cells.
Similarly, we observed ambient ADTs of other markers and scar can remove these ambient signals but preserve ‘true’ ADTs.
[11]:
for marker in ["CD3", "CD4", "CD8a", "CD11b", "PD-1", "IgG2a_control"]:
tmp = ADTs_stacked[ADTs_stacked['ADTs']==marker]
plt.figure(figsize=(3, 1.8))
g = sns.boxplot(x="subtype",
y="log2(ADT+1)",
data=tmp,
hue="count",
hue_order=['raw ADT', 'scar-denoised ADT'],
orient="v",
dodge=True,
width=0.8,
showfliers=False,
linewidth=0.5,
);
g.legend_.set_title('')
g.set(xlabel='', title=f'{marker} protein counts');
g.set_xticklabels(g.get_xticklabels(),rotation=45, ha='right')






UMAP of ADT and RNA#
match ADT name and transcript name
[12]:
prot2gene = PBMCs5k.uns['prot2gene']
Expression of mRNA
[13]:
mRNA = PBMCs5k[:, list(prot2gene.values())].to_df()
mRNA.columns = mRNA.columns.astype(str)+'_RNA'
[14]:
raw_ADT.columns = 'raw_' + raw_ADT.columns.astype(str)
denoised_ADT.columns = 'denoised_' + denoised_ADT.columns.astype(str)
PBMCs5k.obs = PBMCs5k.obs.join(mRNA).join(raw_ADT).join(denoised_ADT)
[15]:
sc.settings.set_figure_params(dpi = 150, figsize = (1.5, 1.5), fontsize = 6)
for marker in ["CD3", "CD4", "CD8a", "CD19", "CD11b", "PD-1"]:
raw_prot_label = f'raw_{marker}'
denoised_prot_label = f'denoised_{marker}'
RNA_label = f'{prot2gene[marker]}_RNA'
sc.pl.umap(PBMCs5k,
size = 5,
color = [raw_prot_label, denoised_prot_label, RNA_label],
color_map = "viridis",
frameon = False,
vmax = [8, 8, 3],
vmin = 0,
wspace = 0.1
)






